Pathogenic fungi are a prominent threat to global food resources by infecting major crops and causing yield loss. Magnaporthe oryzae is a pathogenic fungus, which is known to cause rice blast disease and also can infect other grasses including millet, barley, and wheat. Recent methods of controlling the fungus by selective crop cultivation have proven to be ineffective, as the fungus can rapidly develop resistances and adaptations to these changes.
Specialized metabolites have many roles in conferring evolutionary advantage in nature, by providing virulence, stress protection, and the production of metal transport agents to the organism. The role of specialized metabolites produced as adaptive tools by these fungal species are not yet clearly understood. Here, we focus on diterpenes, a class of specialized metabolites widely distributed in fungal organisms. Magnaporthe pathotypes contain an expanded array of diterpene synthases that may have key roles in the life cycle of the fungus.
Phylogenetic analysis of previously characterized diterpene synthases suggests that Magnaporthe pathotypes produce manoyl oxide using an enzyme that may have evolved from kaurene-producing enzymes in unrelated Ascomycete species. The focus of this project is to evaluate the molecular underpinnings of diterpene synthase evolution in Magnaporthe. This will help answer questions on diversification of terpene metabolism in Magnaporthe pathotypes and give insights into horizontal gene transfer and diversification of diterpenes in Ascomycete in general.
My activities through the CAFE program began with a broad literature review on what we know about terpenes synthases on the molecular and enzymatic levels thus far. In gathering this information, building a new hypothesis which expands upon previous experiments is then attainable. I spent most of my time over the summer analyzing these papers, and extracting from them the key points and representing them in a way which would make the direction of our wet lab work more efficient.
I separated this work into three main steps. First, I made an annotated bibliography of the papers I read with written descriptions of some points that stood out to me regarding the experiments performed. After this I made a spreadsheet which detailed the enzymes studied, mutations made, and changes to enzymatic product outcomes that were observed for each paper. Lastly, I made a multiple sequence alignment of the terpene synthases with known sequences in order to compare the key mutation sites from their respective papers to the sequence of the synthase we are studying currently.
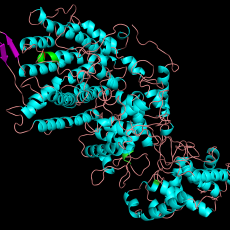
In addition to this literature review, I also used a homology modeling software (Phyre) in order to make a predictive model of our enzyme of study. Being able to then look at this enzyme and its tertiary structure will help give insights into which residues within and surrounding its active sites may play important roles in the class II and class I reaction mechanisms.
Throughout the process of my work over this summer, I have had the opportunity to first get much more experience with effectively reading and analyzing scientific articles to be able to utilize the knowledge gained toward practical purposes in the lab. I feel that this was always something that I wanted to dedicate more time to, but have always felt too busy to do so over the course of my previous semesters. I have also had the chance to utilize a number of bioinformatics tools and techniques such as making multiple sequence alignments, homology models based on protein FASTAs, BLAST searches, and STRING analysis. These tools and techniques have immeasurably helped me get a better understanding of the work that I do in the lab, and its implications to the project at hand.